"Do I have COVID?" This time last year, the question was not in our collective vocabulary. But today, symptoms such as a fever, cough, or headache could prompt a person to wonder: is it COVID-19 or just a cold? Or maybe the flu? These days, this is a very important distinction. Furthermore, based on evidence that people can spread the SARS-CoV-2 virus without showing any symptoms whatsoever, “Am I infectious?” is another important new question to ponder.
Accurate, rapid diagnostic testing prevents disease spread by identifying infectious people who should self-isolate. Testing also helps scientists understand key characteristics of a disease outbreak such as infectivity rate and patterns of spread. There are many challenges, however, to implementing widespread testing for a novel virus such as SARS-CoV-2, and throughout the pandemic, testing availability and reliability have been variable across the United States and around the world. This makes it difficult to contain the virus because when people aren’t aware that they are infected, they are more likely to spread the disease.
Current COVID-19 tests fall into two categories. Lab-based tests that detect the RNA genome of the virus are the most reliable, but they are expensive and require skilled technicians and equipment that are limited in availability. Rapid tests that detect viral proteins are easier to administer and can give results in as little as 15 minutes; however, many are far less accurate than RNA tests. Further, both types of tests could show false negatives if used too early or too late during an infection, when there are fewer virus particles to detect.
Since the beginning of the pandemic, Los Alamos has been addressing these challenges in multiple ways. The Laboratory quickly established a new onsite testing lab and also contributed valuable information to state and federal governments about test efficacy. In addition, Los Alamos programs—built on long-standing capabilities in genomics and protein design—are refining existing tests and also creating new approaches that could improve the future of testing overall.

Testing, testing everywhere
When the novel coronavirus SARS-CoV-2 emerged in late 2019, scientists rapidly determined its full genomic sequence in order to identify exactly what was making people sick. This sequence of about 30,000 ribonucleic acid (RNA) bases is effectively the viral blueprint: it contains all the information necessary to build the virus. The sequence also shows how SARS-CoV-2 is different from other coronaviruses, especially the closely related 2002 SARS virus (SARS-CoV). Even small variations in genetic sequence can translate into major differences in how the virus behaves and why it caused a global pandemic.
Diagnostic tests, however, do not require a full genomic sequence to be determined from each person’s nasopharyngeal swab. Instead, because the genome sequence is known, scientists can identify unique signature regions of the RNA to target in a process called reverse transcriptase polymerase chain reaction (RT-PCR). RT-PCR is considered the gold standard for diagnosing SARS-CoV-2.
Clinical labs often conduct RT-PCR tests, or assays, for diagnostics, but this technology is also widely used in biological research. The presence of PCR machines and skilled personnel in many academic institutions and the DOE national labs meant that additional COVID-19 testing capacity could be added quickly. In April 2020, Los Alamos managers and scientists created an in-house COVID-19 Testing Lab to test Los Alamos staff and to provide support for the New Mexico Department of Health, if needed.
“We were happy to provide this service to the institution,” says Los Alamos Bioscience Deputy Division Leader Alina Deshpande, who manages the COVID-19 Testing Lab. “In fact, our Biological Agent Testing Laboratory (BATL) was already accredited for test evaluation, so it was relatively easy to prepare for the additional certifications.”
Since 2009, the BATL has been evaluating tests based on nucleic acids for various government programs. In other words, Los Alamos would study assays to determine if they accurately detect the pathogen they claim to detect. To do this, the BATL team obtained accreditation from the International Standards Organization, demonstrating its adherence to regimented operations and protocols, such as unidirectional workflows, segregation of workspaces, and strict chains of custody—all to reduce contamination risk.
To process clinical human samples, Deshpande and her team applied for Clinical Laboratory Improvement Amendments (CLIA) registration, which is required for all diagnostic labs. This additional certification required a few steps, such as customizing the information management system and proficiency testing for all staff with blinded samples.
Accurate testing prevents disease spread by identifying infectious people who should self-isolate.
Medical technicians at Los Alamos’s Occupational Medicine group take nasopharyngeal swabs from patients, put them in preservative reagents called viral transport media, and send them to the COVID-19 Testing Lab. There, RT-PCR is done to confirm the presence or absence of SARS-CoV-2 RNA in the sample. Since April, a cohort of about 20 staff members in the Bioscience Division have balanced their normal research priorities alongside taking shifts in the Testing Lab.
The Los Alamos Testing Lab, like others, uses COVID-19 testing kits, reagents, and procedures authorized by the Food and Drug Administration (FDA). The FDA grants Emergency Use Authorizations for specific products and methods as guidance to all clinical labs about what kits are acceptable during the pandemic. To help with this, the FDA and the Centers for Disease Control (CDC) call upon Los Alamos and other national labs to provide independent evaluation of various COVID-19 testing components.
“We are providing data to the FDA and CDC on the effectiveness of different tests and methods to help them make COVID-19 testing decisions at the national level,” explains Bioscience Division Leader Elizabeth Hong-Geller. Hong-Geller is the Los Alamos lead for the R&D lab testing working group within the National Virtual Biotechnology Laboratory, which was established to coordinate COVID-19 research among the Department of Energy’s national laboratory complex. “We have investigated such questions as the stability of nasopharyngeal swabs at different storage temperatures and the reliability of different viral transport media to ensure that RNA on the swabs doesn’t degrade during transport from the clinic to the testing lab.”
Start with the sequence
The first diagnostic tests for COVID-19 were based on the full genomic sequences of SARS-CoV-2 that were available in early 2020. Since that time, scientists around the world have continued to assemble thousands more SARS-CoV-2 sequences for comparison. Most genomes are deposited into online databases such as the National Institute of Health’s GenBank and the Global Initiative on Sharing All Influenza Database (GISAID), which expanded to include SARS-CoV-2. As of today, there are nearly 200,000 genomes available to scientists, which is vitally important for an effective pandemic response.
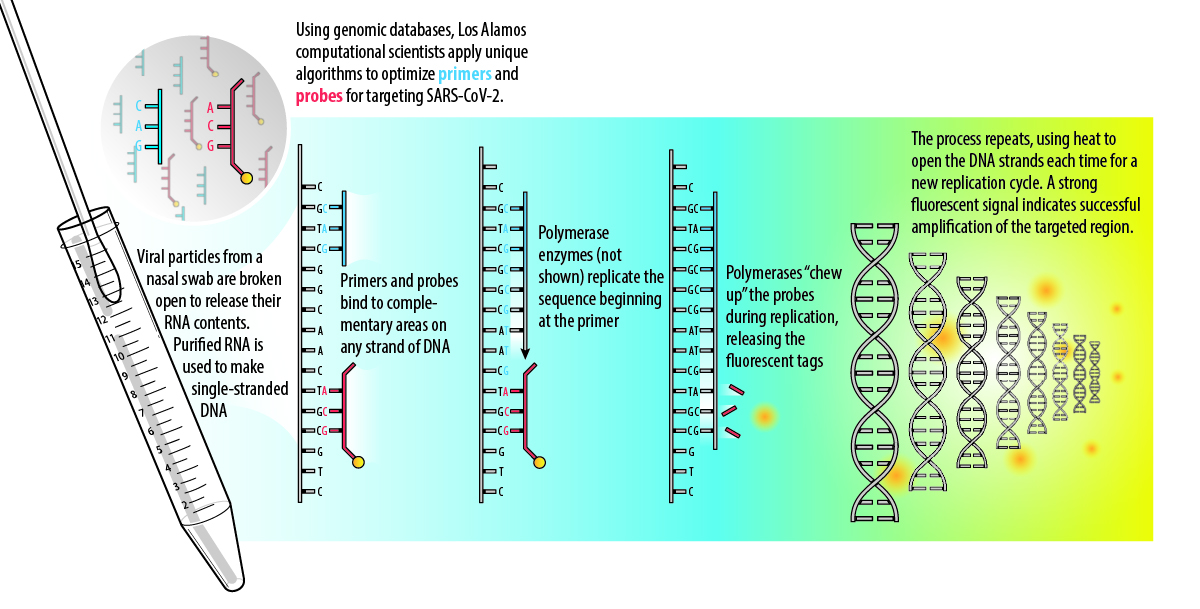
Primers and probes that reliably recognize SARS-CoV-2 are critical to the effectiveness of RT-PCR tests.
In March 2020, Los Alamos bioinformaticist Patrick Chain and his colleagues quickly adapted their award-winning open source software EDGE (Empowering the Development of Genomics Expertise) to help others contribute SARS-CoV-2 genomes to these databases. The EDGE platform provides automated workflows to help users assemble raw genomic data into high-quality complete genomes that are more useful for research. So far, users in more than 50 countries have used COVID-EDGE for their SARS-CoV-2 genomes.
The sequences in these databases help researchers track and study mutations in the SARS-CoV-2 genome to better understand the virus’s phylogeny, or family tree. Because the genetic sequence determines the virus’s structure, the locations of mutation within the genome have important implications. Multiple teams at Los Alamos are evaluating sequence data to track mutations for developing therapeutics and vaccines. Chain’s team is studying current PCR-based tests and has developed an assay-validation tool to determine if tests continue to accurately detect SARS-CoV-2 as its genome mutates.
“This validation can help identify when mutations may interfere with diagnostic tests or even therapeutics. If mutations arise in the RNA regions targeted by an assay, and if these mutations are maintained in the circulating population, it could result in assays no longer being as effective,” says Chain. “Our tools are constantly and automatically being updated to identify where on the planet mutations are arising that make assays more likely to fail.”
Primers, probes, and PCR
The RT-PCR assay is based on some key processes in living cells: complementary binding and nucleic-acid replication. When cells divide to make more cells, they replicate their DNA using enzymes called polymerases. The double-stranded DNA separates into two template strands where the bases are exposed to allow a new copy to be made. The polymerases create a copy strand by assembling new pieces of DNA according to complementary binding with the template: this dictates that the base adenine (A) binds to thymine (T), and guanine (G) binds to cytosine (C). (When an organism has RNA instead of DNA, as in the case of SARS-CoV-2, the enzyme reverse transcriptase is used to make a DNA copy via complementary binding to the RNA template.)
In 1985, scientists developed PCR as a way to mimic this process in a lab setting in order to copy small, undetectable amounts of DNA many times over, amplifying it to a point where it becomes detectable. PCR can also be used as a diagnostic assay by only amplifying signature regions that identify a pathogen. This requires lab-made DNA primers, which determine where along the organism’s DNA strand the replication process will begin, and probes (also made of DNA), which produce a signal—such as a distinctive fluorescent glow—to indicate when amplification of the target has happened. For this reason, the design of primers and probes that reliably recognize SARS-CoV-2 is critical to the effectiveness of the assay.
Los Alamos has multiple projects underway that include advanced primer and probe design for SARS-CoV-2. Overall, making primers and probes is a mathematical optimization problem that must take into account all of the known viral genomic diversity—including any mutations—and also satisfy other criteria for the assay to work.
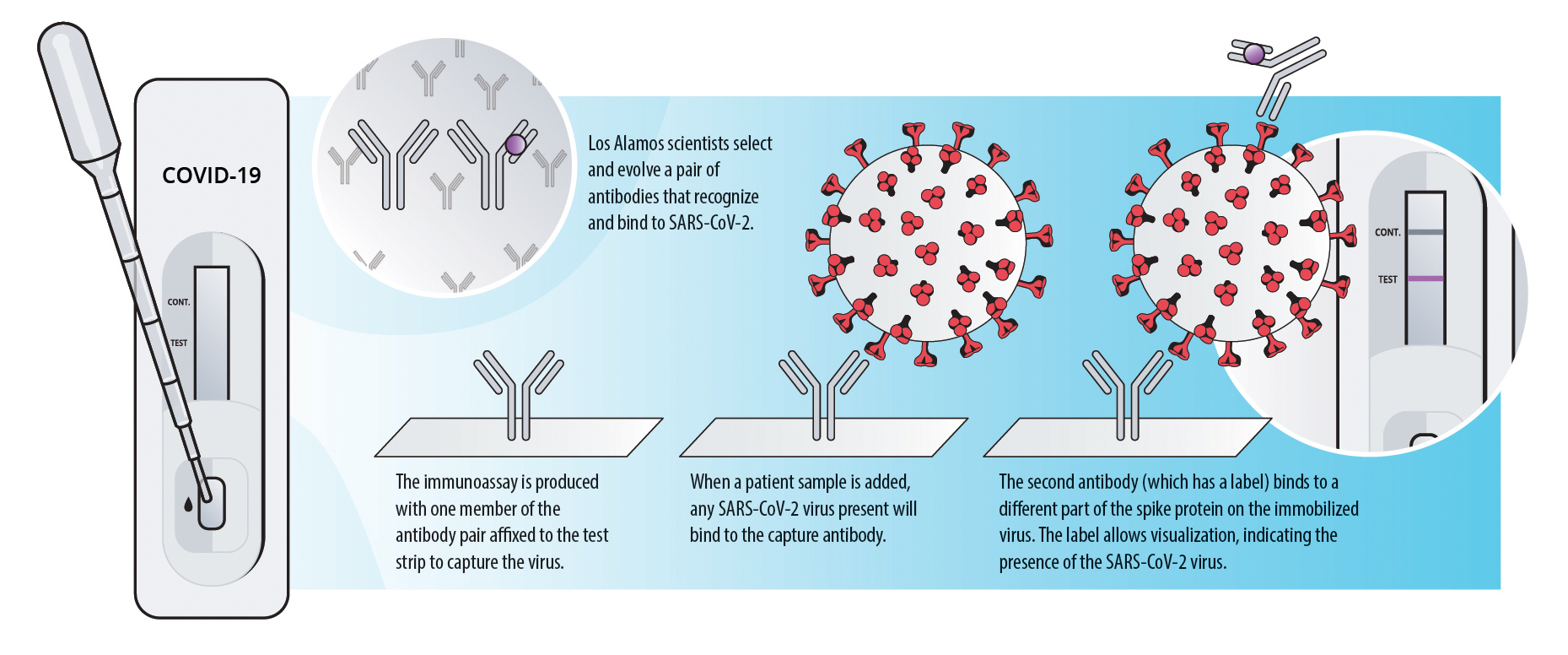
Reliable rapid tests can be developed using highly selective antibodies.
“You need primers and probes that are simultaneously conserved in and unique to the pathogen of interest,” says Jason Gans, a Los Alamos computational biologist. Genes that encode for viral spike proteins are some of the places where mutations have occurred that distinguish SARS-CoV-2 from other closely related viruses. Additional criteria for good primers and probes address size and composition. A good size for probes is about 30 bases; they should include significant amounts of G-C pairs for thermodynamic stability; and they should not inadvertently bind to each other, only to the template.
In one Los Alamos project, Gans and his colleagues are optimizing probes for multiplex assays that detect multiple pathogens at once. This would be especially useful during flu season when there are many viruses in circulation. Gans and his colleagues optimized primers and probes for each target that will not bind to each other or cross-compete for binding sites when they are used at the same time. Furthermore, each probe requires a fluorescent tag, so the team is working to reduce background fluorescence from the multiple probes to ensure a clear signal. So far, Gans says they have designed an assay that simultaneously detects three pathogens and are now aiming for up to sixteen.
Gans is also working with another, larger team at Los Alamos to develop probes for a more portable kind of diagnostic test—one that would still be RNA-based but would eliminate the need for a PCR machine. Biochemist Jessica Kubicek-Sutherland and theoretical biologist Karina Yusim lead a multidisciplinary team that has adapted a project they already had underway—called Fast Evaluation of Variable Emerging Risks (FEVER). The FEVER project first focused on designing probes and instrumentation to support both diagnostics and global surveillance of influenza viruses. When SARS-CoV-2 emerged, the team began to use their FEVER strategy to optimize new coronavirus probes.
“We developed two probes that react with all SARS-like viruses, including SARS-CoV-2,” says Kubicek-Sutherland. These probes are useful for surveilling genomes sampled from various animals to look for newly emerging coronaviruses. “We also developed two probes that only detect SARS-CoV-2 and do not react with any other SARS-like viruses,” she continues. Together, these four coronavirus probes comprise an assay that can be used for both specific SARS-CoV-2 diagnostics and universal coronavirus surveillance, which is important for identifying the next potential pandemic threat.
The team is currently validating its probes in clinical samples, and it is also examining probe sensitivity using saliva, which would be less invasive than nasopharyngeal swabs and does not require a trained technician. The use of saliva is one important aspect of a more portable diagnostic; the other is simplified instrumentation.
The ultimate goal is to adapt these probes to a format that does not require PCR amplification. This would significantly reduce the amount of time and cost it takes to get a test result. Kubicek-Sutherland is part of a Los Alamos team led by Harshini Mukundan that has spent years designing a waveguide biosensor that uses a laser to identify molecules for pathogen detection and is currently adapting the biosensor to be more portable. The team envisions a future with miniature versions of this technology—paired with FEVER probes—which together could eliminate the need for lengthy lab tests by making RNA detection more accessible.
Take-home testing
Waiting several days for a COVID-19 test result makes it difficult to contain the pandemic. For this reason, scientists worldwide have developed—and are improving—rapid tests, many of which use antibodies to detect viral proteins. Suitable inexpensive antibody-based technologies, called immunoassays, have existed for many years. The key ingredient to making immunoassays into a reliable COVID-19 test is highly selective antibodies that only target SARS-CoV-2 and strongly bind to it.
Diagnostic tests can also use this antibody-antigen relationship to detect active, current infections. However, although the antibodies produced by a human during an infection are effective enough to summon an immune response, they are not particularly reliable for diagnostics because one antibody may recognize multiple organisms. If the antibodies in a test recognize many similar coronaviruses, the test could give a false positive result. On the other hand, if the antibody doesn’t bind to enough virus particles, it could give a false negative.
Los Alamos biologists Geoff Waldo, Mietta Lillo, Nileena Velappan, Ramesh Jha, and Hau Nguyen, who comprise the Los Alamos Affinity Reagent Team (LA-ART), are using their long-standing capability to create pairs of highly specific antibodies that work in concert to target SARS-CoV-2 exclusively. The goal is to find two antibodies that will not compete for the same binding site on a target antigen so they can be used in an immunoassay where one antibody captures the virus and the second one creates a detectable signal. Other immunoassays use this approach, but often the target antigen is the SARS-CoV-2 nucleocapsid (a structural protein associated with the viral RNA); however, the Los Alamos team has chosen a different antigen.
“Our target antigen is a component of the SARS-CoV-2 spike protein called the receptor binding domain, or RBD,” says Lillo. “The RBD is a key part of the spike protein that helps it attach to ACE2 receptors in the human body during infection. RBD is also the part of the spike protein where SARS-CoV-2 differs the most from the 2002 SARS virus.”
The LA-ART scientists’ approach uses microorganisms to produce and display human antibodies on their surfaces. These antibodies have the potential to recognize multiple coronaviruses, but the team is able to experimentally focus only on the antibodies that bind most selectively to the SARS-CoV-2 RBD target. The scientists select the microorganisms whose displayed antibodies bind to RBD and allow those microorganisms to reproduce. They repeat this process through multiple generations, evolving the microorganisms to produce antibodies with higher specificity, affinity, and robustness.
Furthermore, by isolating the microorganisms that produce the best-binding antibodies, the team is also able to obtain the genetic code for those antibodies, which will allow the scientists to improve the antibody-antigen interaction through modeling. “Using protein-modeling software, we are able to improve the affinity and change the binding specificity of some of the selected antibodies,” says Los Alamos computational biologist Ramesh Jha. Jha and others also study the molecular dynamics of the protein-protein interactions, which helps them design other novel affinity molecules, similar to antibodies, from scratch.
The invisible enemy
The virus is everywhere, yet it is invisible. It has infected millions, killed hundreds of thousands of Americans, and left others with very few symptoms—and testing is the only way to find it. The nation and the world have come together to tackle this problem. Using research-laboratory assets and a wealth of sequence data, testing facilities and kits were quickly established to diagnose as many people as possible. But the pandemic is still growing, and for as long as people are getting sick, testing will be a major part of trying to contain it. By applying robust scientific approaches, Los Alamos scientists are designing the next generation of tests to help us control this invisible threat. LDRD
Tracking the Invisible Threat
Detecting virus particles once they leave their human hosts is the focus of another National Virtual Biotechnology Laboratory project—Viral Fate and Transport—which is led by Pacific Northwest National Laboratory and Lawrence Berkeley National Laboratory. Large droplets produced from coughing or sneezing are known to fall out of the air quickly. However, smaller ones (less than 100 microns, or millionths of a meter) that are produced when people speak, sing, or shout can stay aloft in poorly ventilated spaces. Determining the impact of these small airborne droplets, called aerosols, is of utmost importance for keeping people safe.
As part of this large project, Los Alamos atmospheric chemist and aerosol specialist Allison Aiken is contributing her team’s expertise and instrumentation at the Los Alamos Center for Aerosol Forensics and Experiments (CAFE). Aiken explains that more experimental data are needed to understand aerosols under different environmental conditions and more realistic circumstances. For example, many previous experiments used simple saline solutions to simulate SARS-CoV-2-containing droplets, which are in reality more complex.
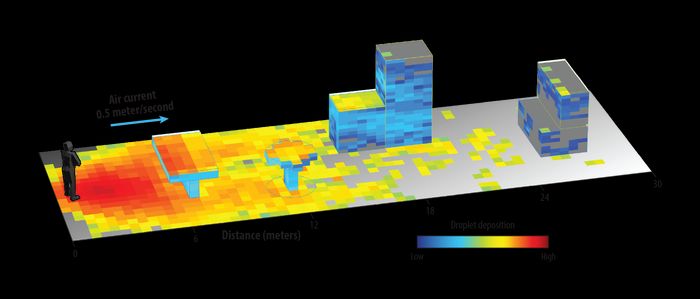
This output shows that a light breeze can transport evaporating
100-micron droplets more than 18 meters away from the infected
individual and deposit them on various surfaces.
“Our team is investigating other surrogates that include proteins and organics in water droplets, in addition to saline, that can result in phase separations under different conditions,” says Aiken. “Using these surrogate liquids, we can create a burst of different-sized droplets in our mixing chambers to study how various temperatures and humidities affect droplet evaporation. This is important because viral fate under low humidity is currently unknown for SARS-CoV-2.”
Using Aiken’s experimental data, Los Alamos atmospheric scientist Michael Brown is using the Quick Urban and Industrial Complex (QUIC) droplet dispersion model to simulate how these various-sized droplets distribute in realistic spaces such as a restaurant courtyard.
“We can change the shape and geometry of the courtyard, adjust spacing of tables and furniture, add or remove overhead canopies, or modify the height of the walls to determine how to make it safer,” says Brown. “We can even add potted bushes and trees to see how they modify the airflow and potentially filter out and catch the virus-filled droplets.”
The Fate and Transport collaboration is also working on other fronts, such as detecting SARS-CoV-2 in wastewater. Wastewater monitoring could be used as an early warning system for future outbreaks—and sequencing wastewater-collected genomes is key. Los Alamos’s genomic scientist Armand Dichosa is contributing to this effort by collaborating with wastewater plants on standardizing sample collection and preparation to get the most useful sequence data. Ultimately, by further understanding the fate and transport of SARS-CoV-2 in multiple environments, Los Alamos scientists hope to provide data to support improved public health guidelines.